A Synthetic Molecular 819 Knot
‘Braiding a molecular knot with eight crossings’ Jonathan J. Danon, Anneke Krüger, David A. Leigh, Jean-François Lemonnier, Alexander J. Stephens, Iñigo J. Vitorica-Yrezabal and Steffen L. Woltering, Science, 355, 159-162 (2017). Full Article.

An 819 knot [Image credit: Stuart Jantzen, www.biocinematics.com]
As every fisherman and sailor knows, different types of knots have different characteristics that make them more or less suited for a particular task: ‘bend knots’ give the strongest binding between two lengths of rope; ‘hitches’ are best for tying rope around an object; and ‘loop knots’, or ‘nooses’, allow degrees of movement between the components they connect.1 There is no reason to believe that different types of knots won’t be just as important, versatile and useful in the molecular world. But we can’t find out about that until we can make them.
Knots occur in DNA and approximately 1% of proteins,2 and they form spontaneously in polymer chains3 (just as happens with plates of spaghetti and headphone cables!). However, until now synthetic chemists have only been able to make the simplest types of molecular knot,4 such as trefoil5 (three crossings) and pentafoil6 (five crossing) knots. This is partly because the most widely used methods to make molecular knots twist two ligand strands around one another.4 The unavailability of all but the simplest molecular knots hinders the investigation of the effects of different knot types in materials and other molecular applications.
Now scientists at the University of Manchester have developed a way of braiding three molecular strands enabling tighter and more complex knots to be made than has previously been possible.7 Ligand I is heated with FeCl2 in N,N-dimethylformamide (DMF) (Step 1, Figure 1). At 130 °C the iron ions reversibly attach to and detach from different strands until the lowest energy structure is formed, a cyclic triple helix II which is stabilized by a chloride ion bound at its center through C-H···Cl- hydrogen bonds and Fe2+-Cl- electrostatic interactions. A catalyst is then employed to join the end groups of II to form the closed loop 819 knot III (Step 2, Figure 1). Removal of the iron ions from III with NaOH (Step 3, Figure 1) forms the wholly organic 819 knot IV.
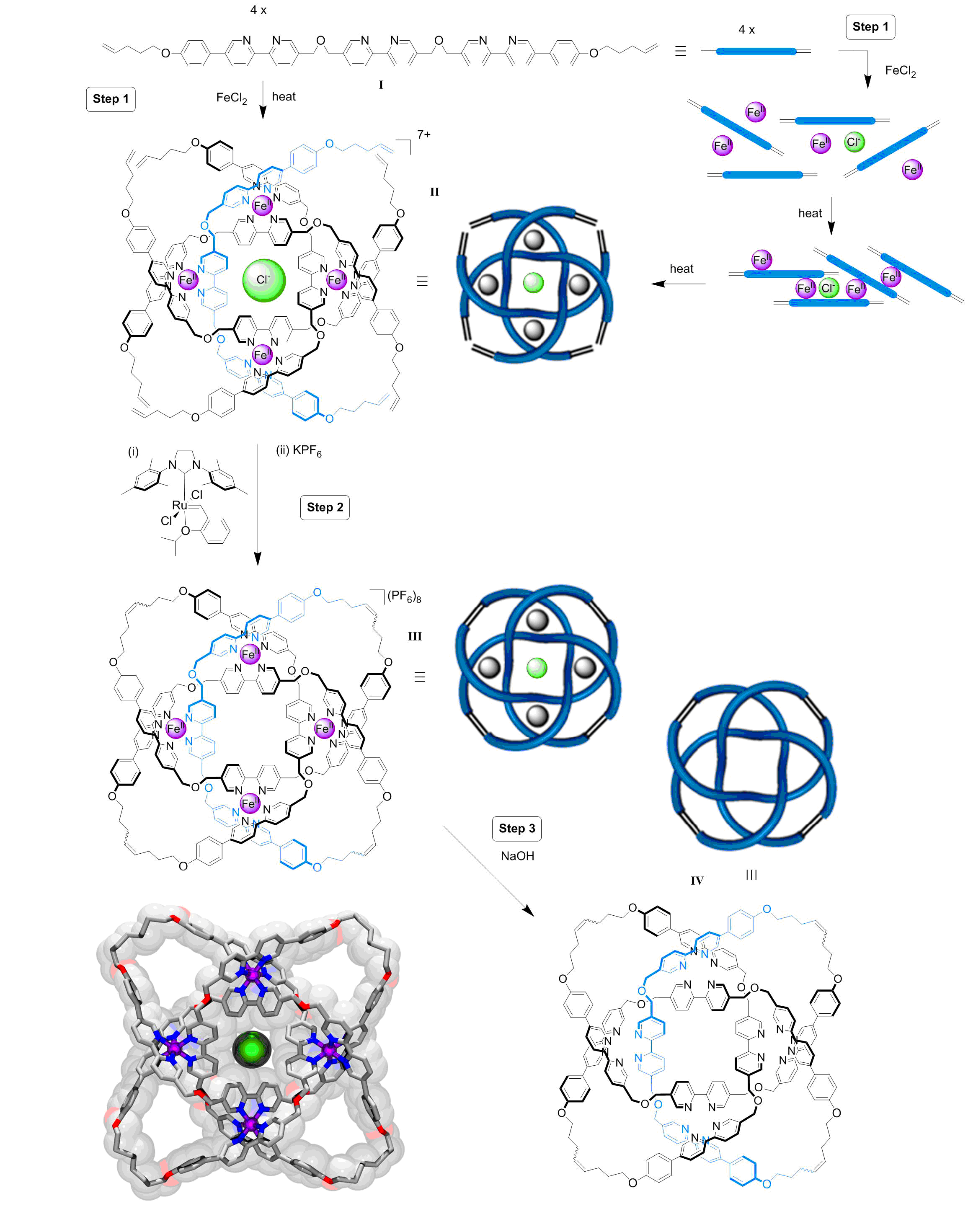
Figure 1. It’s a kind of magic: Assembly of a molecular 819 knot [video credit: Stuart Jantzen, www.biocinematics.com].
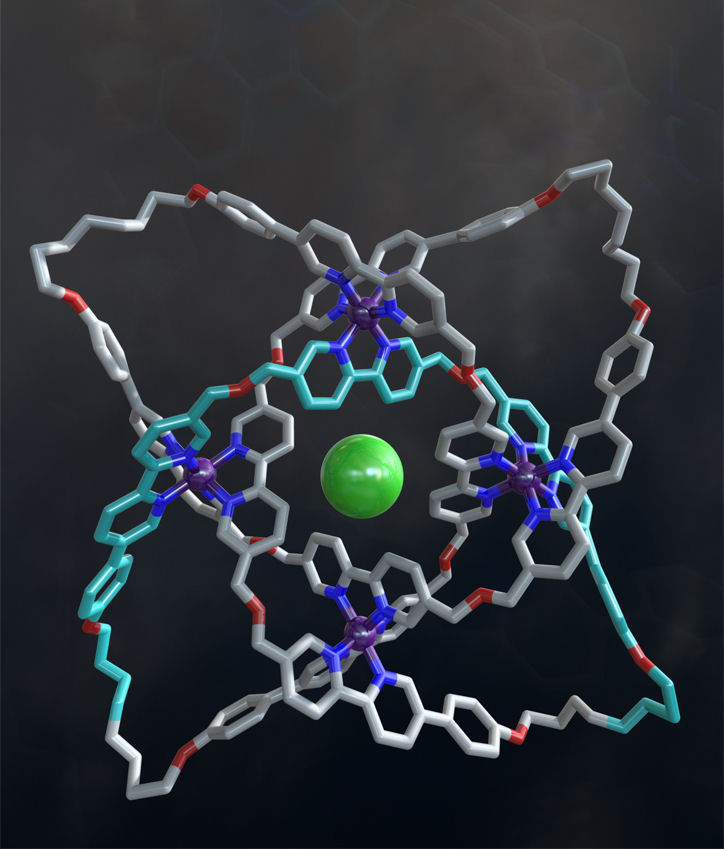
The approach was used to make a molecular knot consisting of a 192-atom (20 nm) continuous loop interwoven with eight non-alternating crossings (Figure 2). It is the mostly tightly knotted physical structure known at ~2.5 nm (24 atoms) per crossing. Unusually, the molecular knot can be made in right or left handed forms solely by virtue of its topology (the knot possesses no elements of Euclidean chirality – stereogenic centers such as asymmetric atoms, helical subunits, etc. which are usually found in molecules of single handedness).
Figure 2. A molecular knot with eight crossings. The X-ray crystal structure of a 192-atom-loop molecular 819 knot featuring iron ions (shown in purple), oxygen atoms (red), nitrogen atoms (dark blue), carbon atoms (shown in metallic grey, with one of the building blocks shown in light blue) and a single chloride ion (green) at the centre of the structure. Image credit: Robert W. McGregor (www.mcgregorfineart.com).
Being able to make different types of molecular knots means that scientists should be able to probe how knotting affects strength and elasticity of materials and may enable them to weave polymer strands to generate new types of material.
The first synthetic molecular knot, a trefoil knot, was made by Jean-Pierre Sauvage’s group in 1989.5 For related synthetic strategies applied in the field of artificial molecular machines Sauvage shared the 2016 Nobel Prize in Chemistry.8 More than six billion different prime knots are known to mathematics;9 in the quarter of a century since Sauvage’s breakthrough chemists have been able to make just four different types of knot.
The synthesis of the molecular 819 knot illustrates a strategy (the braiding of three strands) that should be applicable to the synthesis of many more, and more diverse, types of molecular knots. By making different types of molecular knot we can find out about their properties (such as at which point are entangled molecular strands prone to breaking?) and which knots have properties best suited for a particular purpose, just as a fisherman or sailor knows which knot is best suited for each task.

An 819 knot [Image credit: Stuart Jantzen, www.biocinematics.com]
References
[1] D. Pawson ‘Handbook of Knots: Expanded Edition’, Dorling Kindersley Ltd, London, 2004.
[2] N. C. H. Lim and S. E. Jackson. ‘Molecular knots in biology and chemistry’, J. Phys.: Condens. Matter 27, 354101 (2015).
[3] M. D. Frank-Kamenetskii, A. V. Lukashin and A. V. Vologodskii ‘Statistical mechanics and topology of polymer chains’, Nature 258, 398-402 (1975).
[4] J.-F. Ayme, J. E. Beves, C. J. Campbell and D. A. Leigh ‘Template synthesis of molecular knots’, Chem. Soc. Rev. 42, 1700-1712 (2013).
[5] C. O. Dietrich-Buchecker and J.-P. Sauvage ‘A synthetic molecular trefoil knot’, Angew. Chem., Int. Ed. Engl. 28, 189-192 (1989).
[6] (a) J.-F. Ayme, J. E. Beves, D. A. Leigh, R. T. McBurney, K. Rissanen and D. Schultz ‘A synthetic molecular pentafoil knot’, Nature Chem. 4, 15-20 (2012).
.....(b) V. Marcos, A. J. Stephens, J. Jaramillo-Garcia, A. L. Nussbaumer, S. L. Woltering, A. Valero, J.-F. Lemonnier, I. J. Vitorica-Yrezabal and D. A. Leigh. ‘Allosteric initiation and regulation of catalysis with a molecular knot’, Science 352, 1555-1559 (2016).
[7] J. J. Danon, A. Krüger, D. A. Leigh, J.-F. Lemonnier, A. J. Stephens, I. J. Vitorica-Yrezabal and S. L. Woltering. ‘Braiding a molecular knot with eight crossings’, Science 355, 159-162 (2017).
[8] ‘Jean-Pierre Sauvage - Facts’, Nobelprize.org. Nobel Media AB 2014. Web. 8 Jan 2017.
[9] E. E. Fenlon ‘Open problems in chemical topology’, Eur. J. Org. Chem. , 5023-5035 (2008).