A Read-Only Molecular Turing Machine
‘A Tape-Reading Molecular Ratchet’ Yansong Ren, Romain Jamagne, Daniel J. Tetlow, David A. Leigh, Nature, 612, 78–82 (2022).. Full Article.
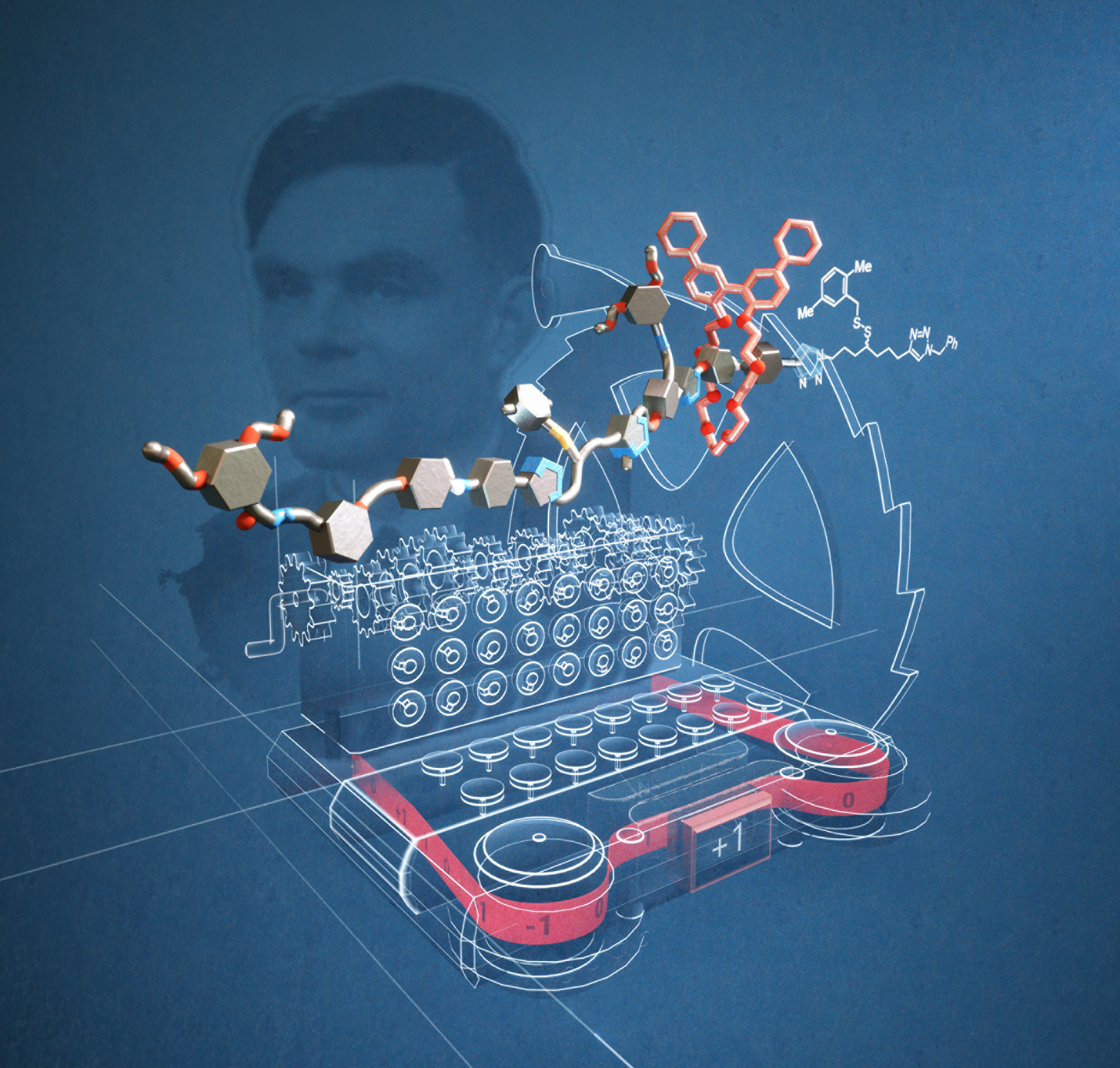
In 1936 Alan Turing published a concept for an automatic machine for abstract computing.1 Nowadays called a ‘Turing machine’, his proposal envisaged a device that featured a ‘head' that could read and write symbols while moving along a tape. This influential thought experiment helped spark the development of modern computer science. Although no practically useful device that functions according to Turing’s original design has ever been made (modern computers use a different design that employs random access memory), the resemblance of Turing’s ‘automatic machine’ to information processing in biology is striking (the reading and writing of information by the ribosome, for example2,3). Now scientists at the University of Manchester have made an artificial molecular machine that moves along a molecular tape, changing the reading head’s shape (conformation) in response to ‘symbols’ on the tape it encounters along the way.4 The geometrical change alters how light is twisted (clockwise, anticlockwise, or not twisted) by the head and so the sequence of symbols on the molecular tape can be read out as a string of data according to the changing twisting of light (clockwise corresponds to the digit +1; anticlockwise to -1, not twisted to 0).
The new nanomachine is a type of machine termed a ‘finite state automaton’2: that is, a Turing machine that moves in one direction through a string-encoded state sequence, with readable outputs (here the changing twist in the light emitted) but is not (yet) able to write symbols itself (which would be a full Turing machine). The machine’s invention opens the way for the reading—and ultimately writing—of information using the powered directional movement of artificial nanomachines along molecular tapes.
The molecular machine features a crown ether that is pumped from solution onto a molecular tape by a pulse of chemical fuel.5 Further fuel pulses transport the macrocycle through a series of compartments of the thread, via an energy ratchet mechanism,6 before releasing the macrocycle back to the bulk solution off the other end of the tape (Fig. 1). During its directional transport along the tape the crown ether changes conformation according to the stereochemistry of binding sites it encounters along the way.7 This allows sequence information programmed into the tape to be read out as a string of digits in a non-destructive manner through a changing circular dichroism (CD) response (Fig. 2).8 The fuelling synchronizes dynamics so that almost every macrocycle is at the same tape position simultaneously, causing a large and measurable response of the ensemble. The concept was exemplified by the reading of molecular tapes with strings of balanced ternary digits (‘trits’9) -1,0,+1 and -1,0,-1. An animation of the operation of the tape-reading molecular machine is shown in Video 1.
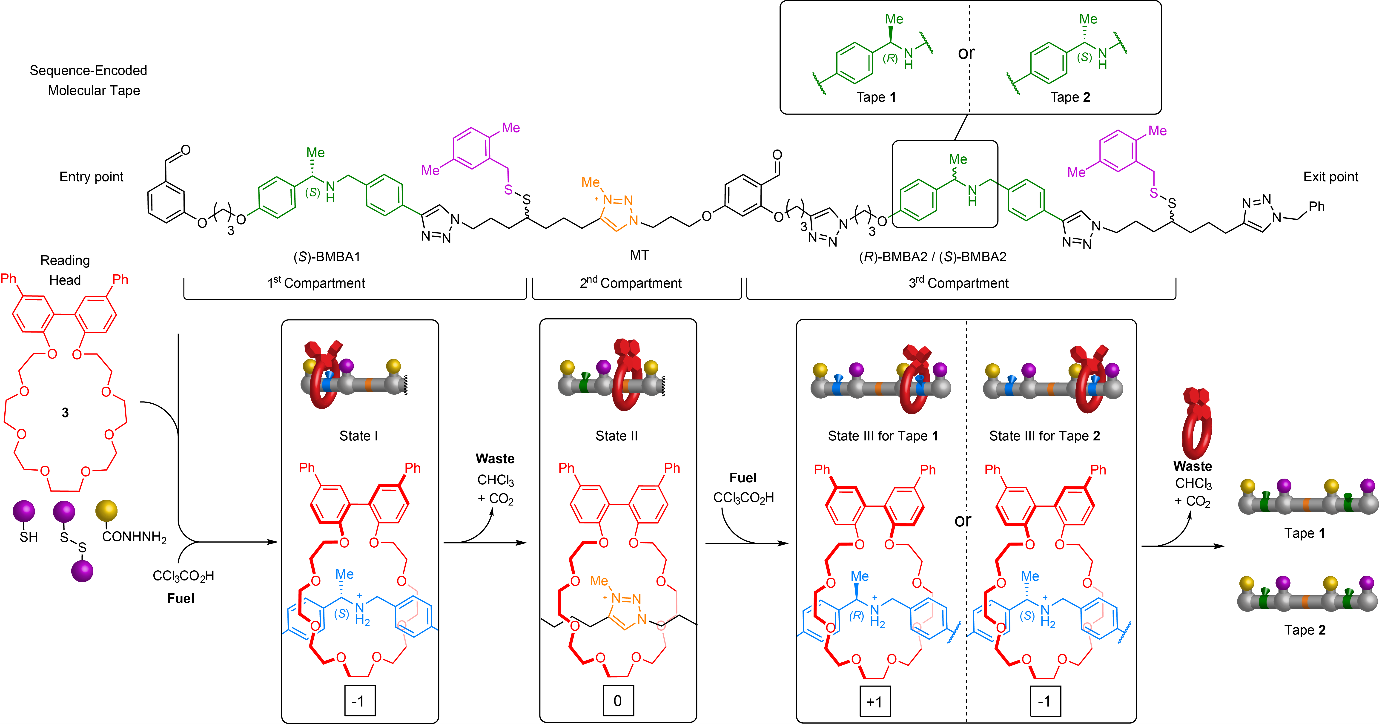
Figure 1: Chiroptical read-out of the sequence of a stereochemically encoded molecular tape by a chemically fueled molecular ratchet. Molecular tape 1 features an (S)-N-benzyl-α-methylbenzylamine ((S)-BMBA·H+) binding site for crown ether 3 in the first compartment, an N-methyltriazolium (MT) binding site in the second compartment, and an (R)-BMBA·H+ binding site in the third compartment. Tape 2 differs from 1 only in the stereochemistry of the binding site in the third compartment ((S)-BMBA·H+). The tape compartments are separated by acid-labile, base-locked hydrazone groups on one side and base-labile, acid-locked disulfide groups on the other.5a Crown ether 3 threads through the entry point at the left terminus of the tape under acidic conditions (e.g. upon addition of CCl3CO2H) and is transported through the compartments from left-to-right in response to oscillations in the pKa of the medium, exiting the tape to the right. As the crown ether passes through the different compartments it changes conformation according to the binding site stereochemistry. The different conformations of the crown ether exhibit different circular dichroism responses. By monitoring the changing CD spectrum the sequence of binding site stereochemistries on the tape is read non-destructively by the macrocycle. Tape 1 is encoded -1,0,+1; tape 2 is encoded -1,0,-1. The molecular ratchet is a finite state automaton, a machine that runs through a string-encoded state sequence, giving outputs dependent on the occupied state.
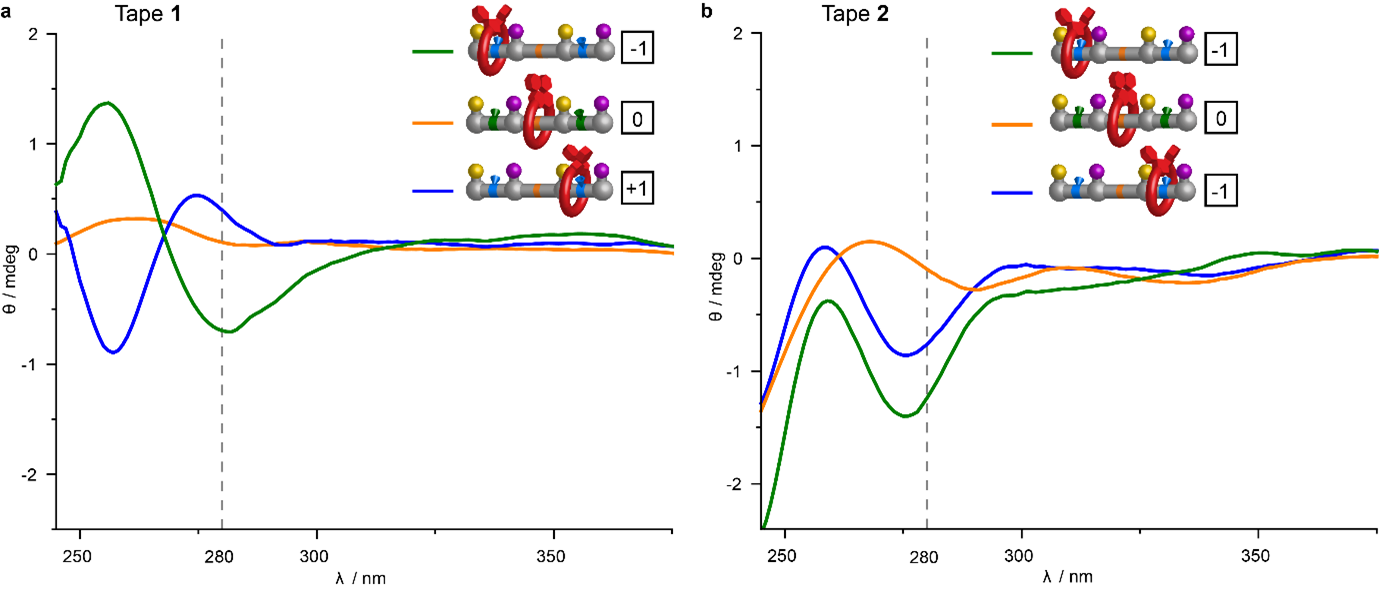
Figure 2: Circular dichroism (CD) spectra of the machine states formed by the stepwise ratcheted operation of macrocycle 3 along molecular tapes 1 (encoded −1,0,+1) and 2 (encoded −1,0,−1). a, CD spectra of components and machine states from the reading of tape 1. b, CD spectra of track and machine states from the reading of tape 2. The machine state can be read by the sign and intensity of the CD spectrum at 280 nm. Experiments were carried out in CHCl3, [C] = 100 μM.
There are a number of particularly noteworthy features about the new system:
(i) In the artificial molecular machine design, stereochemistry is used for the ‘symbols’ to encode information in the molecular tape. This is very different to biology, which uses chemoselectivity (different hydrogen bonding motifs of different nucleotides) for this purpose.2
(ii) Ratcheting6d with a chemical fuel5a is crucial for providing directionality to the reading head. It is the combination of machine components performing different functions10 (ratcheting to providing directional transport of the head, and the reading of stereochemical information from the tape through non-covalent interactions that cause a conformational change in the head) that enables directional reading of information from the molecular tape.
(iii) The use of a pulsed chemical fuel5 synchronises the dynamics so that many of the machines in the ensemble are at the same position on tapes at the same time. This produces a large collective response of the ensemble, which is read non-destructively by circular dichroism (CD) spectroscopy. This is different to the ribosome: individual ribosomes act independently and are not synchronised in terms of their operation with other machines.
(iv) This tape-reading molecular ratchet is the first non-biological example of a molecular finite automaton2; that is a Turing machine that only moves in one direction and reads, but is unable to write, symbols. However, there is clearly a future possibility of using the conformational change in the head to write stereochemical information through catalysis. That would represent a type of Turing machine called a ‘string transducer’2, which is a complete Turing machine except that it only moves in one direction. Protein translation is an example of programing into a string transducer (the ribosome).
The new system marks a major step towards the goal of artificial molecular Turing machines, in which information can be read, translated and written by chemically fuelled molecular machines that move directionally along information-encoded molecular tapes.

Members of the tape-reading ratchet team, Alan Turing Building, University of Manchester, 17th October 2022. Left-to-right: Romain Jamagne, Dr Daniel Tetlow, Prof. David Leigh and Dr Yansong Ren.
References
1. Turing, A. M. On computable numbers, with an application to the Entscheidungsproblem. Proc. Lond. Math. Soc. 42, 230–265 (1936).
2. Benenson, Y. Biomolecular computing systems: principles, progress and potential. Nat. Rev. Genet. 13, 455–468 (2012).
3. Varghese, S., Elemans, J. A. A. W., Rowan, A. E. & Nolte, R. J. M. Molecular computing: paths to chemical Turing machines. Chem. Sci. 6, 6050–6058 (2015).
4. Ren, Y., Jamagne, R., Tetlow, D. J. & Leigh, D. A. A tape-reading molecular ratchet, Nature, 612, 78–82 (2022).
5. (a) Erbas-Cakmak, S., Fielden, S. D. P., Karaca, U., Leigh, D. A., McTernan, C. T., Tetlow, D. J. & Wilson, M. R. Rotary and linear molecular motors driven by pulses of a chemical fuel. Science 358, 340–343 (2017). (b) Biagini, C. & Di Stefano, S. Abiotic chemical fuels for the operation of molecular machines. Angew. Chem. Int. Ed. 59, 8344–8354 (2020). (c) Olivieri, E., Gasch, B., Quintard, G., Naubron, J.-V. & Quintard. A. Dissipative acid-fueled reprogrammable supramolecular materials. ACS Appl. Mater. Interfaces 14, 24720–24728 (2022).
6. (a) Astumian, R. D. & Derényi, I. Fluctuation driven transport and models of molecular motors and pumps. Eur. Biophys. J. 27, 474–489 (1998). (b) Hernández, J. V., Kay, E. R. & Leigh, D. A. A reversible synthetic rotary molecular motor. Science 306, 1532–1537 (2004). (c) Chatterjee, M. N., Kay, E. R. & Leigh, D. A. Beyond switches: Ratcheting a particle energetically uphill with a compartmentalized molecular machine. J. Am. Chem. Soc. 128, 4058–4073 (2006). (d) Kay, E. R., Leigh, D. A. & Zerbetto, F. Synthetic molecular motors and mechanical machines, Angew. Chem. Int. Ed. 46, 72–191 (2007).
7. Kuwahara, S., Chamura, R., Tsuchiya, S., Ikeda, M. & Habata, Y. Chirality transcription and amplification by [2]pseudorotaxanes. Chem. Commun. 49, 2186–2188 (2013).
8. (a) Bottari, G., Leigh, D. A. & Pérez, E. M. Chiroptical switching in a bistable molecular shuttle. J. Am. Chem. Soc. 125, 13360–13361 (2003). (b) David, A. H. G., Casares, R., Cuerva, J. M., Campaña, A. G. & Blanco, V. A [2]rotaxane-based circularly polarized luminescence switch. J. Am. Chem. Soc. 141, 18064−18074 (2019).
9. Davis, M., Sigal, R. & Weyuker, E. J. Computability, Complexity, and Languages and Logic: Fundamentals of Theoretical Computer Science (2nd Edn.). San Diego: Academic Press, Harcourt, Brace & Co. (1994).
10. Zhang, L., Marcos, V. & Leigh, D. A. Molecular machines with bio-inspired mechanisms. Proc. Natl. Acad. Sci. USA 115, 9397−9404 (2018).
